Transforming biology to design next-generation computers, using a surprise ingredient

Moore's law—which says the number of components that could be etched onto the surface of a silicon wafer would double every two years—has been the subject of recent debate. The quicker pace of computing advancements in the past decade have led some experts to say Moore's law, the brainchild of Intel co-founder Gordon Moore in the 1960s, no longer applies. Particularly of concern, next-generation computing devices require features smaller than 10 nanometers—driving unsustainable increases in fabrication costs.
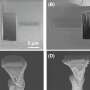
Biology creates features at sub-10nm scales routinely, but they are often structured in ways that are not useful for applications like computing. A Purdue University group has found ways of transforming structures that occur naturally in cell membranes to create other architectures, like parallel 1nm-wide line segments, more applicable to computing.
Inspired by biological cell membranes, Purdue researchers in the Claridge Research Group have developed surfaces that act as molecular-scale blueprints for unpacking and aligning nanoscale components for next-generation computers. The secret ingredient? Water, in tiny amounts.
"Biology has an amazing tool kit for embedding chemical information in a surface," said Shelley Claridge, a recently tenured faculty member in chemistry and biomedical engineering at Purdue, who leads a group of nanomaterials researchers. "What we're finding is that these instructions can become even more powerful in nonbiological settings, where water is scarce."
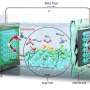
In work just published in Chem, sister journal to Cell, the group has found that stripes of lipids can unpack and order flexible gold nanowires with diameters of just 2 nm, over areas corresponding to many millions of molecules in the template surface.
"The real surprise was the importance of water," Claridge said. "Your body is mostly water, so the molecules in your cell membranes depend on it to function. Even after we transform the membrane structure in a way that's very nonbiological and dry it out, these molecules can pull enough water out of dry winter air to do their job."
Their work aligns with Purdue's Giant Leaps celebration, celebrating the global advancements in sustainability as part of Purdue's 150th anniversary. Sustainability is one of the four themes of the yearlong celebration's Ideas Festival, designed to showcase Purdue as an intellectual center solving real-world issues.
The research team is working with the Purdue Research Foundation Office of Technology Commercialization to patent their work. They are looking for partners for continued research and to take the technology to market.
More information: Ashlin G. Porter et al, 1-nm-Wide Hydrated Dipole Arrays Regulate AuNW Assembly on Striped Monolayers in Nonpolar Solvent, Chem (2019). DOI: 10.1016/j.chempr.2019.07.002