Understanding metabolic processes through machine learning
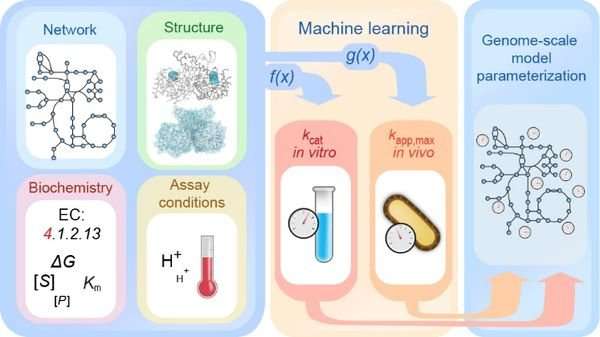
Bioinformatics researchers at Heinrich Heine University Düsseldorf (HHU) and the University of California at San Diego (UCSD) are using machine learning techniques to better understand enzyme kinetics and thus also complex metabolic processes. The team led by first author Dr. David Heckmann has described its results in the current issue of the journal Nature Communications.
The synthetic life sciences rely on a detailed and quantitative understanding of the complex systems in biological cells. Only if such systems are understood is their targeted manipulation possible. A system already well known is biological metabolism, in which many hundred enzymes are involved. However, a key aspect in this area, namely the individual activity of each enzyme, is only insufficiently understood in quantitative terms.
Together with his colleagues in California and Düsseldorf, Dr. David Heckmann, now in San Diego and former doctoral researcher under Professor Martin Lercher at HHU's Institute for Computational Cell Biology, has chosen a bioinformatics approach to get to the bottom of the properties of enzymes. For this, the researchers are using machine learning, a sub-field of artificial intelligence (AI), which is already used successfully in other areas, e.g. traffic management or automated translation. Machine learning algorithms have in recent years defeated their human opponents at chess, Go, and even poker.
Their approach allowed the researchers to identify important enzyme properties that are deciding factors for their activity. With these results, they can describe the kinetics of a large number of enzymes far more clearly than was possible with previous methods.
"Our model provides a fascinating insight into which enzyme properties influence their activity the most," says Professor Lercher.
"We can use this knowledge to model metabolic processes more precisely and to analyse the interaction of various components in cellular networks," adds Dr. Heckmann.
More information: David Heckmann et al. Machine learning applied to enzyme turnover numbers reveals protein structural correlates and improves metabolic models, Nature Communications (2018). DOI: 10.1038/s41467-018-07652-6
Journal information: Nature Communications
Provided by Universität Düsseldorf